SOM based index
The methodology for the spatial segregation indicator is based on 2 publications ((Cottrell et al. 2017),(Olteanu et al. 2019)).This section reproduces the method of how to calculate the indicator for 81 neurons.
library(sp)
library(rgeos)
library(rgdal)
library(ggplot2)Reading spatial database
CLUSTER<-readOGR('data/CLUSTER.shp')Chess distance for each neuron
o<-as.data.frame(matrix(0,81,81))
colnames(o)<-1:81
t<-as.matrix(cbind(9:1,18:10,27:19,36:28,45:37,54:46,63:55,72:64,81:73))
C<-1
for(j in 1:dim(t)[2]){
for(i in 1:dim(t)[1]){
F<-1
for(l in 1:dim(t)[2]){
for(k in 1:dim(t)[1]){
d<-max(c(abs(i-k),abs(j-l)))
o[F,C]<-d
F<-F+1
}
}
C<-C+1
}
}Geographic distance matrix
centroides<-gCentroid(CLUSTER,byid = TRUE)
dist<-spDists(centroides)
dist<-as.data.frame(dist)
colnames(dist)<-as.numeric(rownames(centroides@coords))+1
rownames(dist)<-as.numeric(rownames(centroides@coords))+1
O<-as.data.frame(matrix(0,length(colnames(dist)),length(colnames(dist))))
colnames(O)<-colnames(dist)
rownames(O)<-colnames(dist)
for(i in 1:dim(O)[1]){
for(j in 1:dim(O)[2]){
Nf<-CLUSTER$NEUR[i]
Nc<-CLUSTER$NEUR[j]
D<-o[Nf,Nc]
O[i,j]<-D
}
}
DGEO<-NULL
for(i in 2:dim(dist)[1]){
for(j in 1:(i-1)){
DGEO<-c(DGEO,dist[i,j])
}
}
DSOM<-NULL
for(i in 2:dim(O)[1]){
for(j in 1:(i-1)){
DSOM<-c(DSOM,O[i,j])
}
}plot(DSOM,DGEO)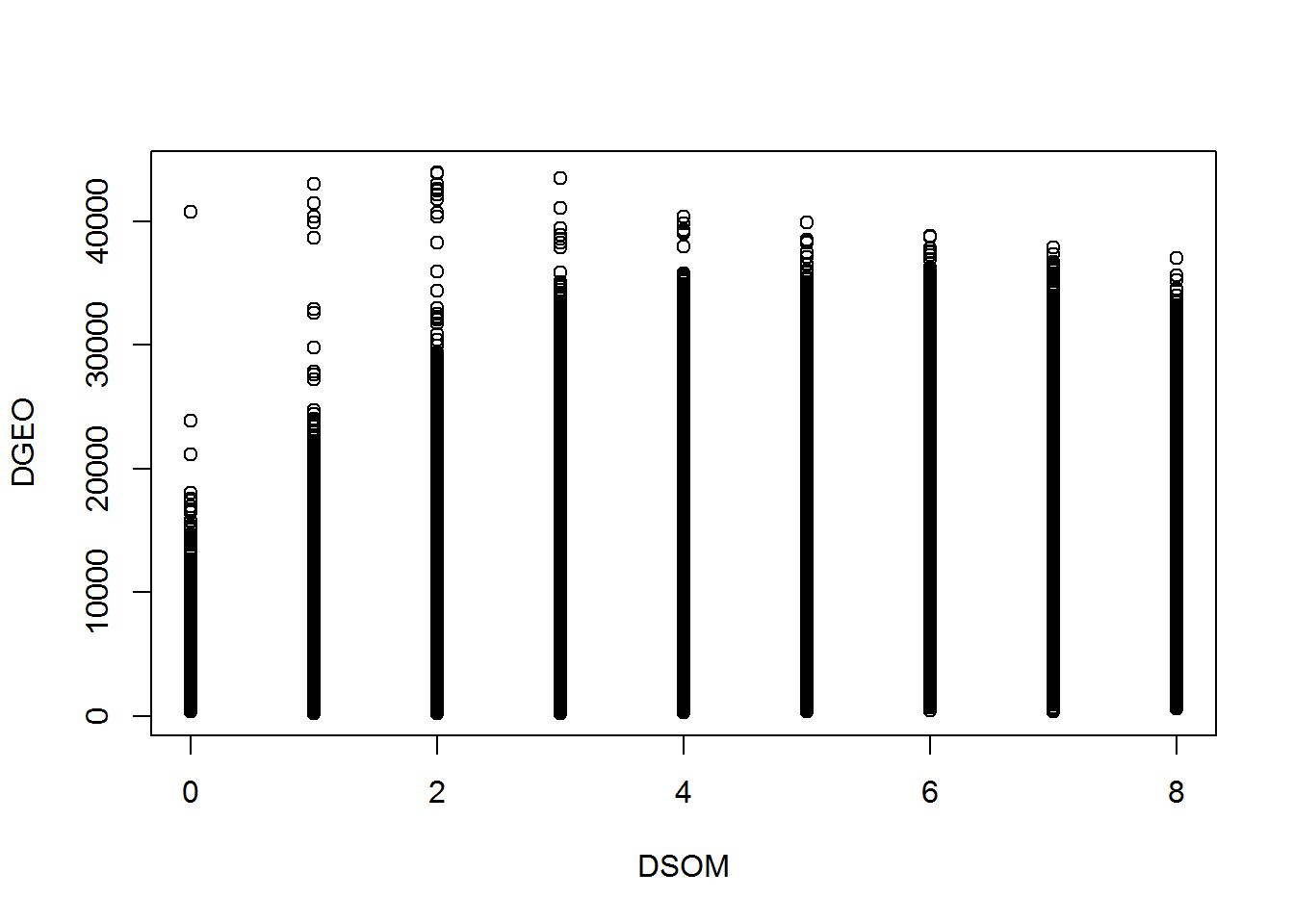
ggplot(data=data.frame(DSOM,DGEO),aes(x=DSOM,y=DGEO)) +
geom_bin2d(bin=20) +
scale_fill_continuous(type = "viridis") +
geom_smooth(method=lm, se=FALSE, fullrange=TRUE)## `geom_smooth()` using formula 'y ~ x'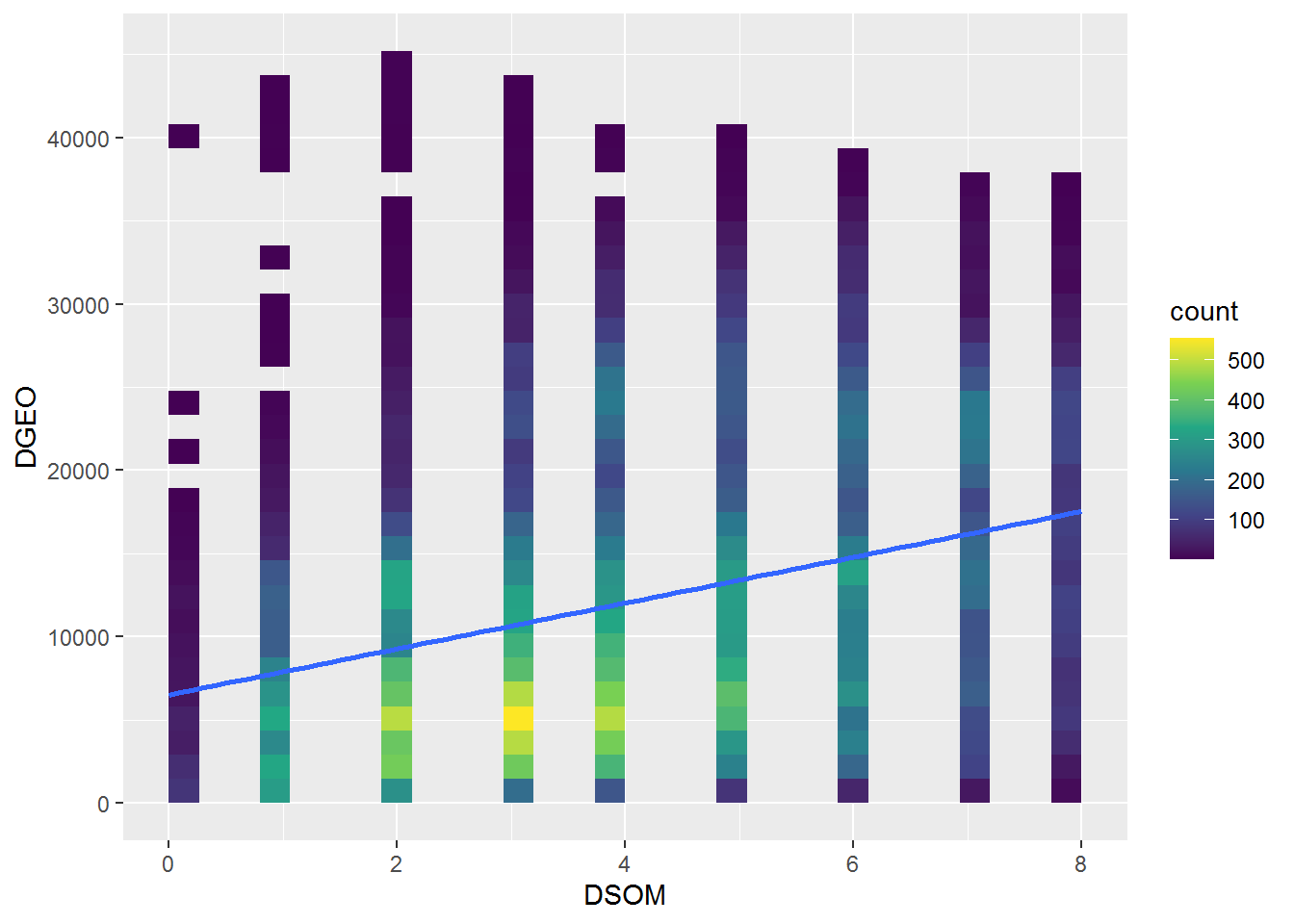
cor(DSOM,DGEO)## [1] 0.3347225References
Cottrell, Marie, Madalina Olteanu, Julien Randon-Furling, and Aurelien Hazan. 2017. “Multidimensional Urban Segregation: An Exploratory Case Study,” 1–7.
Olteanu, Madalina, Aurélien Hazan, Marie Cottrell, and Julien Randon-Furling. 2019. “Multidimensional Urban Segregation: Toward a Neural Network Measure.” Neural Computing and Applications, 1–13.
diegmedina@udec.cl